Protein Origami: Can DeepMind’s AlphaFold revolutionize drug discovery?
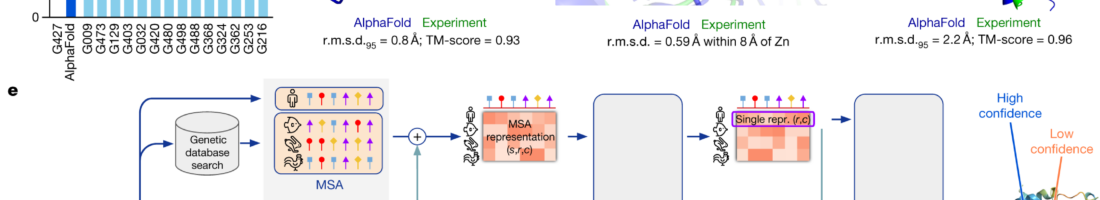
AlphaFold has cracked the code to protein folding, uncovering the shapes of all proteins known to science. How will they or a partner now use this information?
What is a protein and why is it important?
In recent years, mRNA has been the most talked about biochemical molecule, bolstered by the sequencing and open source nature of the human genome and technologies like CRISPR. Much like a genome, all living organisms also have a proteome, which details all of the proteins that can be made by a cell or organism. Proteins in the human body “provide structure, produce energy, as well as allow communication, movement, and reproduction” . For example, proteins in the human body include keratin, which provide the structure of hair and nails, Immunoglobulin G, the most common antibody in blood, and Insulin, a hormone that regulates blood glucose to name just a few.
In effect, proteins are found everywhere in the human body; unfortunately, protein misfolding is “believed to be the primary cause of Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, Creutzfeldt-Jakob disease, cystic fibrosis, Gaucher’s disease and many other degenerative and neurodegenerative disorders.”
The Hard Problem
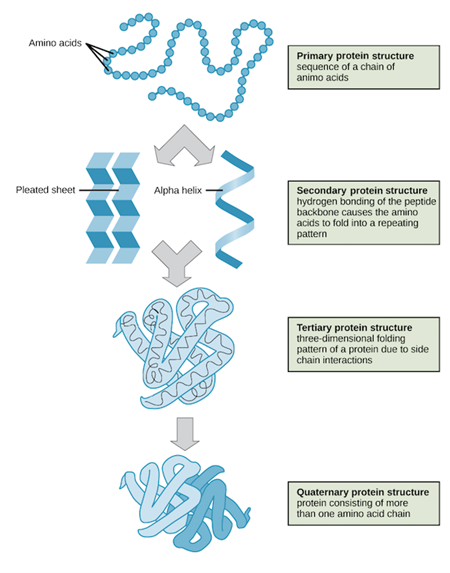
Each protein is comprised of a chain of amino acids (protein building blocks), of which there are 20 unique ones found in most biological life. This chain commonly has 50 individual amino acid beads, and can have as many as 10^300 different configurations . This is due to the myriad combinations of amino acids and their unique chemistry which combine different atomic shapes and create areas of polarity. This is in contrast to nucleic acids, which only have 5 nucleotides (DNA/RNA building blocks), and due to their unique chemistry, always take on the classic double-helix spiral.
The number of combinations alone make protein folding a quantitatively difficult problem to solve, but the method of observing proteins is also a challenge due to their ability to denature (“unfold”) in different environments, and the methods currently available to observe their true forms (X-Ray Crystallography). The non-AI methods were laborious, expensive, and could take years of research. One way to think about it: a blob fish is so named because that’s how it looks out of the ocean depths where it resides. But in the deep ocean, a blob fish looks completely different. This is much the same problem that protein folding faces, in that it is difficult to observe the protein in situ and to understand how and where it functions.
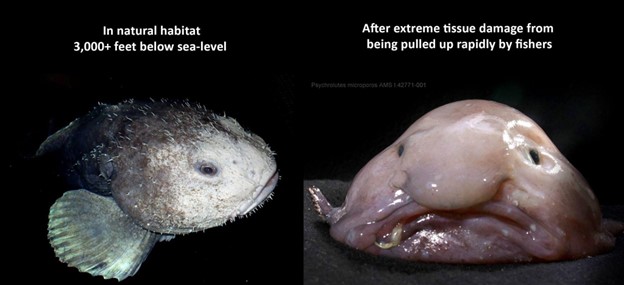
Figure 1: A Picture is worth a thousand words – the blobfish represents the one aspect of the challenge of visualizing proteins under technology constraints.
AlphaFold AI
Deepmind, a subsidiary of Google, set out to solve the hard problem of understanding how all 200 million known proteins fold, using AI in place of traditional scientific tools of measurement.
Timeline:
- Pre 2018: model trained on 100,000 known protein sequences and structures
- 2018: AlphaFold placed first in protein folding prediction competition with record breaking accuracy
- 22 July 2021: AlphaFold publicized 350,000 known proteins including the human proteome
- 28 July 2022: AlphaFold publicized over 200 million structures – nearly every catalogued protein known.
How did they do it?
All of AlphaFold’s methods and specifications are open source and described in their papers published in Nature and other scientific journals. Suffice to say, they are using cutting edge ML technology to reduce the 10^300 combinations for each protein down to just 1, and with extremely high accuracy down to the level of subatomic spacing. In effect, they input an amino acid sequence into their model, and using a neural network, are able to predict which areas of the amino acid chain will fold together and how.

A point solution, or something more?
AlphaFold is completely open source, well documented and transparent. Their product could be seen as two things – their process of how AI predicts how a protein will fold, and there dataset of shapes of known proteins. Both of these products are open source and free to use by any company or institution. Form desktop research, it’s not clear how or if Deepmind operates as a for-profit business line of Google So, it’s ability to capture value lies mostly within who uses AlphaFold’s AI or dataset and their innovations. This method of applying AI in biotech has the potential to work not just as a point solution (solving for how proteins fold) but also as an entire operating model for drug discovery.
Chagas Disease
Currently, AlphaFold is being used to investigate better treatments for Chagas Disease, a parasitic, chronic disease that can lead to heart failure. ALphafold has the complete proteome of the parasite that causes the diseases. Combining the knowledge of the shapes of proteins the parasite makes with all other shapes of known proteins, researchers are able to determine which proteins are most likely to combine and neutralize the parasite’s proteins. They found that their results “provide insight into the mechanisms of action of the compounds and their targets, and pave the way for new strategies to finding novel compounds or optimize already existing ones.”
Potential
It’s unclear if any companies have built on top of the AlphaFold model to use it’s protein folding predictions as a simulator yet, but this is a potential use case that could be implemented in the drug discovery research pipeline. Working in reverse, could scientists use Alphafold to predict novel amino acid chains not already catalogued that would fold into a protein to neutralize an attacking protein? If a person creates an abnormal protein, could Alphafold determine a novel “helper” protein that would rectify the abnormality? Undoubtedly, there are many other predictive processes that could be built on top of AlphaFold’s to find or create new medicines and therapeutics.
Sources
[1] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5054093/
[1]https://pubmed.ncbi.nlm.nih.gov/16689923/#:~:text=Protein%20misfolding%20is%20believed%20to,other%20degenerative%20and%20neurodegenerative%20disorders.
[1] https://www.forbes.com/sites/robtoews/2021/10/03/alphafold-is-the-most-important-achievement-in-ai-ever/?sh=5de341796e0a
[1] https://www.nature.com/articles/s41586-021-03819-2 [1] https://www.frontiersin.org/articles/10.3389/fcimb.2022.944748/full


